- Open sequence files (reference sequence file with features, sequencing data from facility (ABI file with chromatograms))
- Activate reference sequence window.
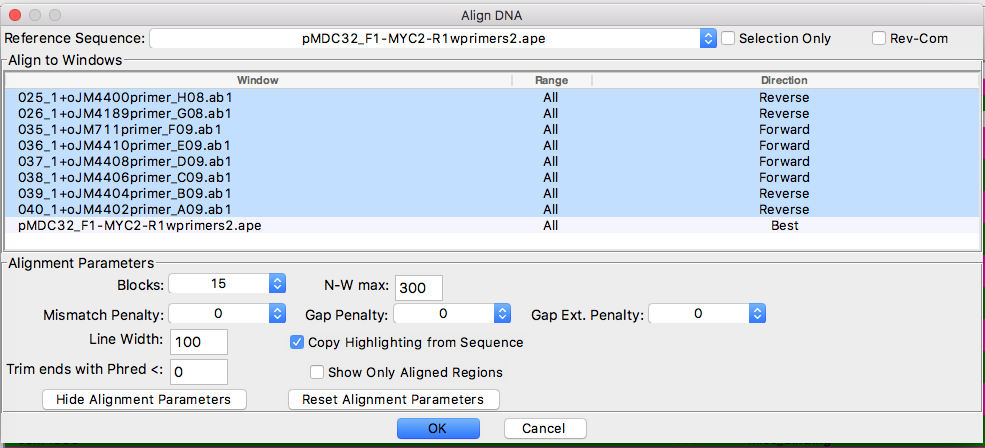
- Tools > Align sequences
- “add” sequences (no need to add reference seq). Note that “Reference Sequence” on top of “Align DNA” window should be reference sequence active.
- determine sequence direction (judged by primer orientation; “BEST” did not work well)
- check “Copy highlighting from sequence”
- click “OK” and save file (automatically as “.rtf”)
- entire insert region should be sequenced twice (both direction), so that mark which region needs to be sequence with which direction and determine primers you need to use for sequencing (or design primers).
## "Align sequences" window
## test result
{width=1200px}